Research
Bacteria behaving badly
The DiRita lab investigates biology and pathogenicity of bacteria associated with
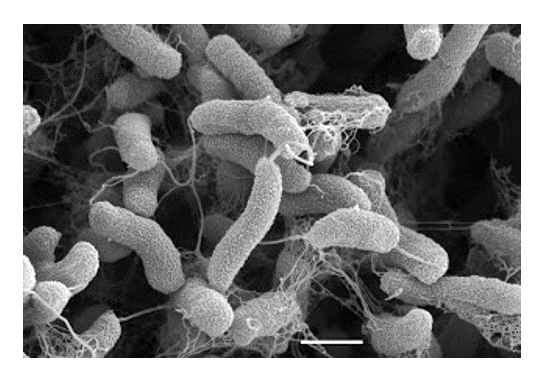 infectious diseases. Vibrio cholerae, the agent of human cholera, expresses two major pathogenicity determinants the cholera
toxin and the toxin-coregulated pilus. We study regulatory proteins that work in concert
to control the expression of these pathogenicity determinants and thereby control
virulence of the microbe. An unusual aspect of these proteins is their membrane localization,
and how transcription regulators engage the transcription complex while remaining
in the membrane opens our research into areas of very basic interest. We have also
investigated the determinants for recognition and uptake of V. cholerae by intestinal M cells, a key aspect of the early immune response against this pathogen.
infectious diseases. Vibrio cholerae, the agent of human cholera, expresses two major pathogenicity determinants the cholera
toxin and the toxin-coregulated pilus. We study regulatory proteins that work in concert
to control the expression of these pathogenicity determinants and thereby control
virulence of the microbe. An unusual aspect of these proteins is their membrane localization,
and how transcription regulators engage the transcription complex while remaining
in the membrane opens our research into areas of very basic interest. We have also
investigated the determinants for recognition and uptake of V. cholerae by intestinal M cells, a key aspect of the early immune response against this pathogen.
Another microbe we investigate is Campylobacter jejuni, one of the most prevalent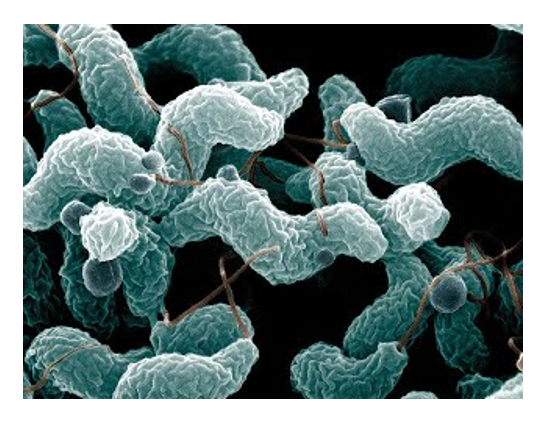 foodborne pathogens in the United States. C. jejuni is a common, benign inhabitant of the chicken gut microbiota, so human infection
is often linked to contaminated chicken meat. It is also generating significant resistance
to antibiotics. We study basic mechanisms underlying the biology and pathogenicity
of C. jejuni, and have developed many tools for genetic analysis of this microbe. We seek to understand
how it behaves during benign association with the chicken and virulent association
with humans (and suitable animal models.) Also, we discovered the major pathway by
which Campylobacter species take up DNA in a process called natural transformation,
and are working to uncover further knowledge about that mechanism.
foodborne pathogens in the United States. C. jejuni is a common, benign inhabitant of the chicken gut microbiota, so human infection
is often linked to contaminated chicken meat. It is also generating significant resistance
to antibiotics. We study basic mechanisms underlying the biology and pathogenicity
of C. jejuni, and have developed many tools for genetic analysis of this microbe. We seek to understand
how it behaves during benign association with the chicken and virulent association
with humans (and suitable animal models.) Also, we discovered the major pathway by
which Campylobacter species take up DNA in a process called natural transformation,
and are working to uncover further knowledge about that mechanism.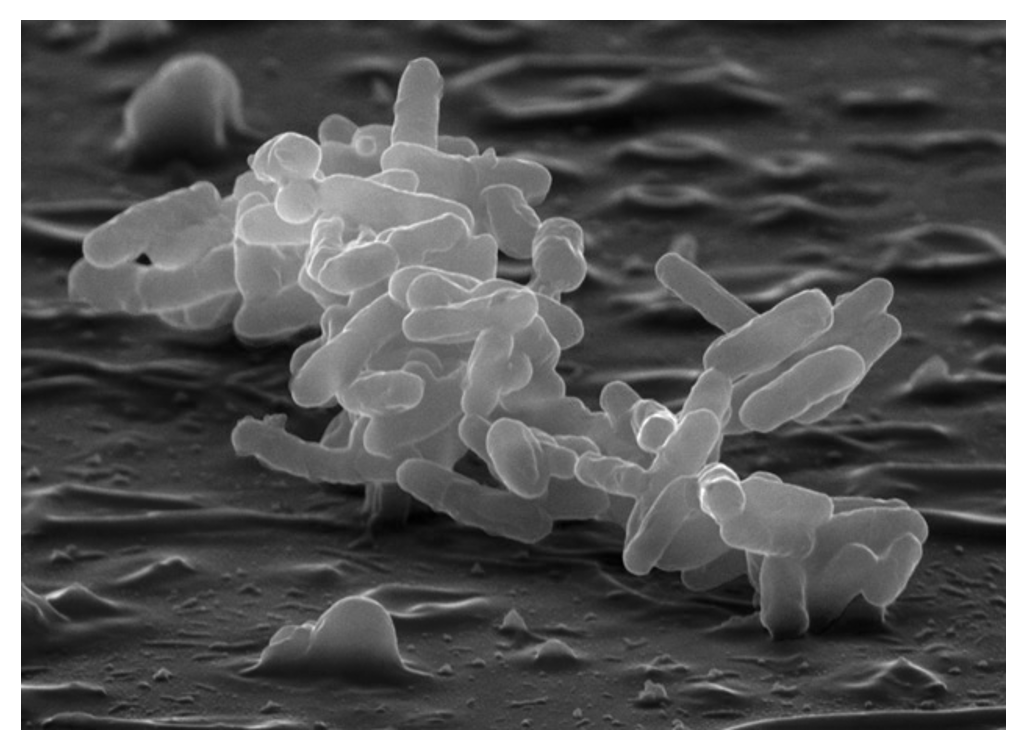 We are in a consortium of investigators studying hospital-acquired microbes of the
so-called “Carbapenem-resistant Enterobacteriaceae” (CRE). Enterobacter cloacae causes serious infections in health-care settings and has acquired resistance to
many different antibiotics. Our work with this microbe includes generating a sequence-arrayed
transposon insertion library to carry out genetic analyses of its colonization and
pathogenicity traits, thereby identifying potential new therapeutic targets to combat
this important opportunistic pathogen.
We are in a consortium of investigators studying hospital-acquired microbes of the
so-called “Carbapenem-resistant Enterobacteriaceae” (CRE). Enterobacter cloacae causes serious infections in health-care settings and has acquired resistance to
many different antibiotics. Our work with this microbe includes generating a sequence-arrayed
transposon insertion library to carry out genetic analyses of its colonization and
pathogenicity traits, thereby identifying potential new therapeutic targets to combat
this important opportunistic pathogen.